The complexity and variability of biomedical language pose a challenge in fields dealing with biomedical data. A vast array of terms, referring to various entities, including living organisms, biochemicals, drugs, biological pathways, diseases, and disease symptoms in patients, is utilized in biomedical research and closely associated healthcare industries. The intricacy inherent in this domain necessitates both a structured representation of knowledge and the use of a standardized language to describe these entities. Medical ontology serves as a valuable tool for fulfilling this crucial purpose.

What is Ontology?
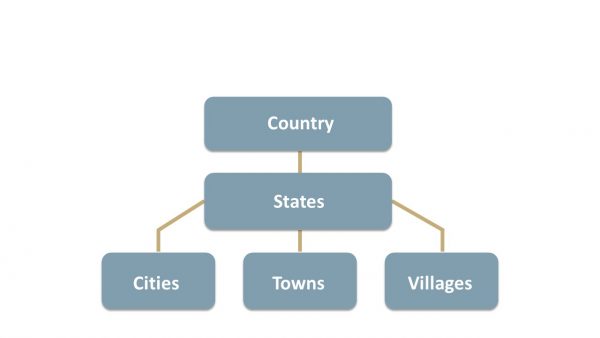
Originally a philosophical study of existence, ontology, in the context of information science, refers to a formal representation of knowledge or information within a specific domain. Ontology provides a structured framework for organizing entities, their properties, and relationships. To illustrate, let’s consider the ontology of a country. As shown below, it encompasses entities such as states, cities, towns, and villages, along with their hierarchy and relationships. Interestingly, a similar ontology focused on geographical information serves as the foundation for the widely used Geographic Information System (GIS).
By extension, medical data is also highly complex—perhaps even more so—due to the vast diversity of biological entities and their interdependent relationships and interactions. This inherent complexity has prompted the development of numerous types of ontologies, which are designed to maintain consistency and accuracy in medical data, records, and research. At present, there are hundreds of medical ontologies, and you can find many of them by following this link.
There are numerous advantages to utilizing ontologies. For biomedical researchers, the use of medical ontologies can assist them in annotating and categorizing data with standardized terminology. This, in turn, leads to clear communication and makes it easier to identify relationships, patterns, and other elements within the data. For medical practitioners, ontologies facilitate the exchange of patient information among healthcare providers and entities, ensuring that data sent and received by different systems are interpreted consistently. This not only enhances the accuracy and consistency of medical information but also promotes interoperability between different healthcare systems. Furthermore, it supports the development of intelligent systems for medical diagnosis and treatment.
Apart from the benefits in research and practice, medical ontology also plays a crucial role in text mining. There is a constant influx of biomedical text from various sources every day. However, due to the intricacies of biomedical concepts and the inherent ambiguity in natural language, precise data extraction from large and heterogeneous data sources is not feasible without a system of standardization. Medical ontology helps mitigate this ambiguity and variability in text data. Furthermore, medical ontology is also pivotal in the Semantic Web, where web data are represented through structured ontological representations, allowing interpretation by both humans and machines. In short, the use of medical ontology leads to more intelligent search, enhanced data integration, and improved knowledge discovery across a wide range of medical and healthcare-related resources.
An overview of commonly used medical ontologies
There are a number of ontologies that are particularly helpful when building text analysis solutions for the medical field. In what follows, we provide an overview of some of the more commonly used ontologies.
MeSH
The Medical Subject Headings (MeSH) dictionary is a hierarchical collection of medical terms describing various conditions, diseases, and symptoms. It was created by the US National Library of Medicine (NLM) at the National Institutes of Health (NIH) in the United States.
MeSH is utilized for indexing and searching biomedical information found in national medical databases, including MEDLINE/PubMed. Many biomedical articles are now indexed with MeSH terms, either manually or automatically by computer, enabling users to retrieve records on a specific subject with accuracy and efficiency.

The MeSH tree structure comprises 16 main branches, such as Anatomy, Organisms, Diseases, and more. Each branch is further divided into hierarchical levels, ranging from more general to more specific categories. These levels represent the taxonomic relationships between terms, as follows:
- synonyms: terms with the same, or nearly the same, meaning. (e.g., “dwelling” and “residence” are synonyms of “home”)
- hypernyms: terms with a broad meaning for a group of specific items. (e.g., “animal” is a hypernym for “dog” and “cat”)
- hyponyms: terms with a more specific meaning than a general term. (e.g., “apple”, “banana”, and “orange” are hyponyms of “fruit”)
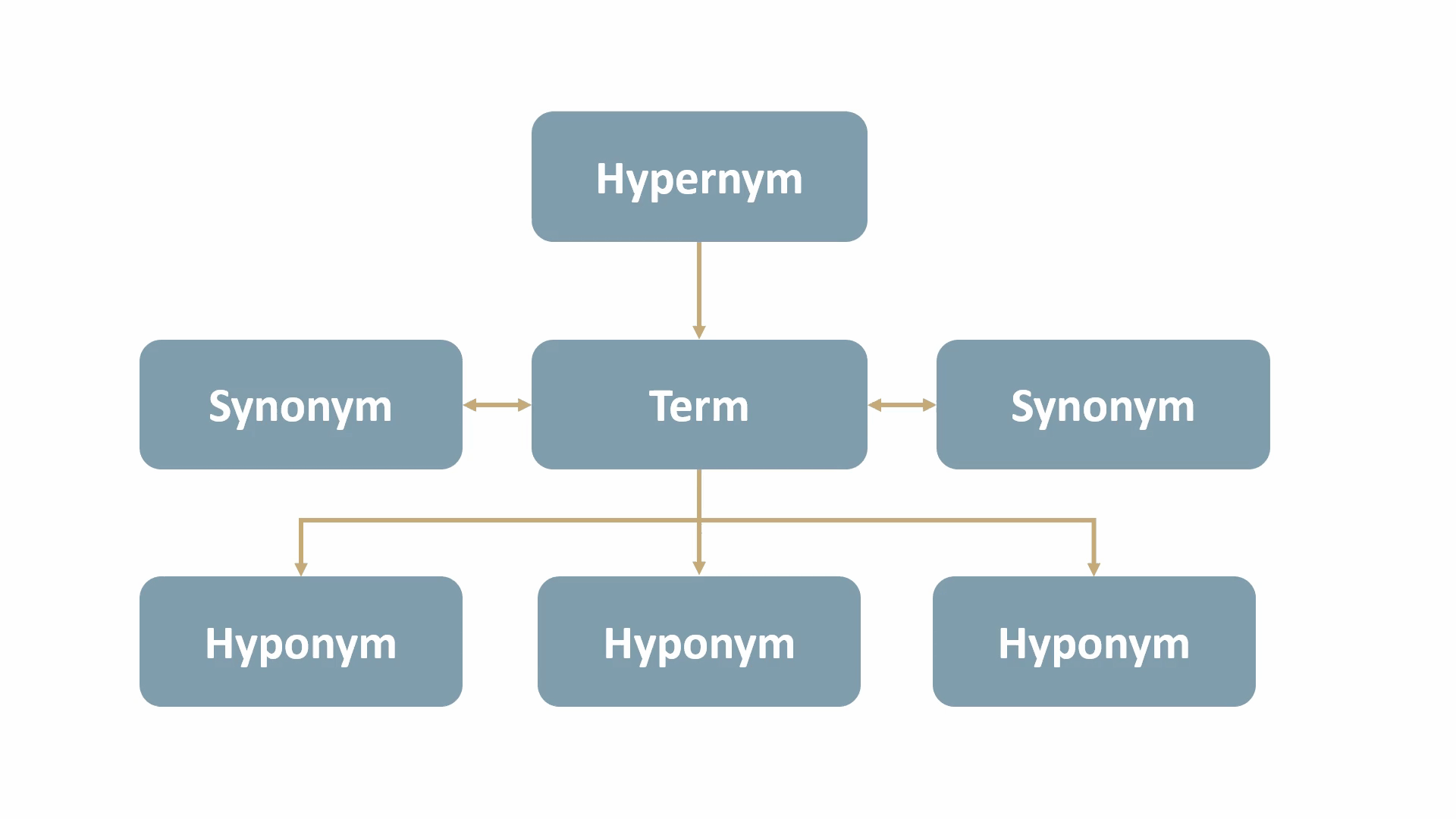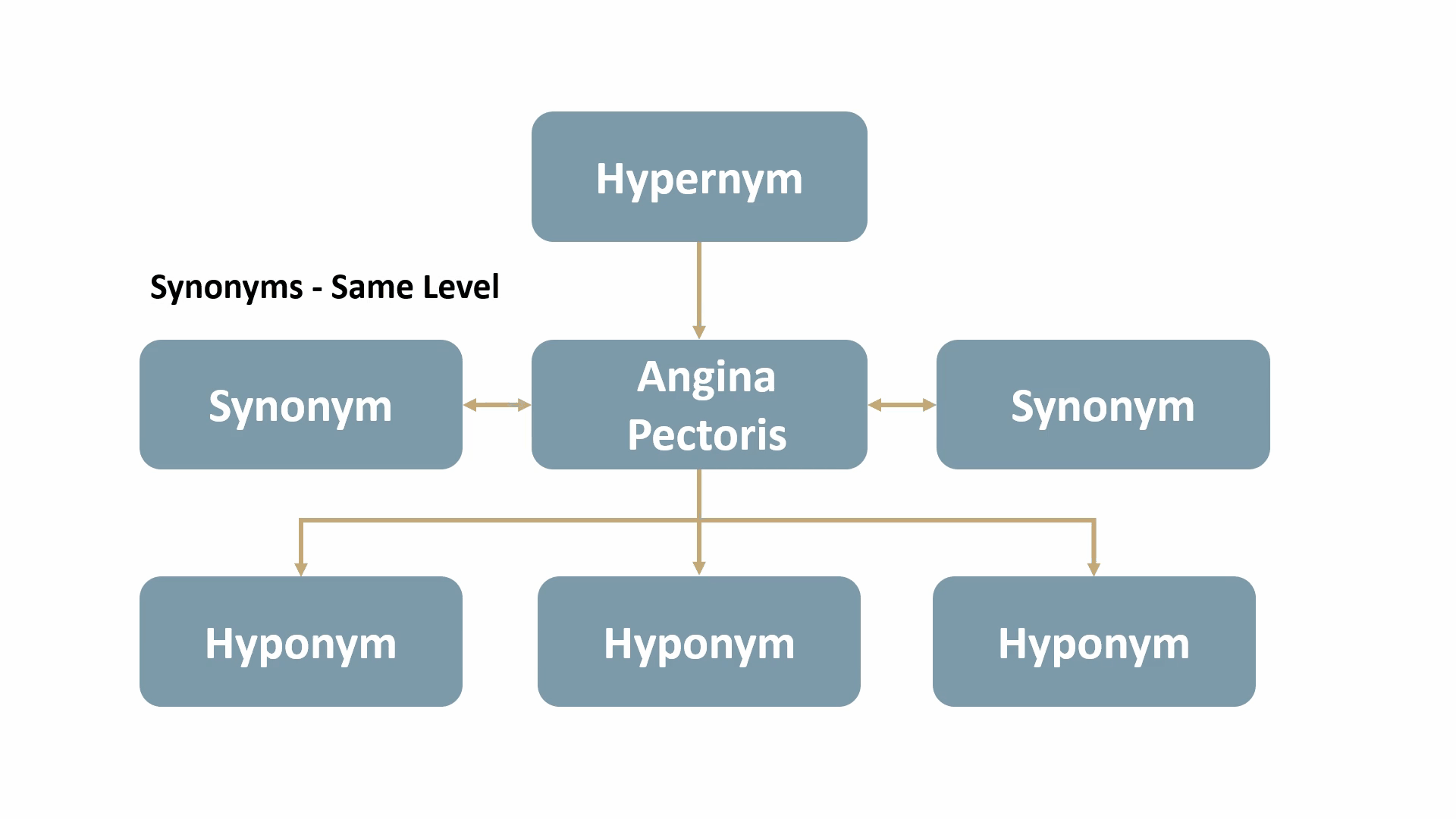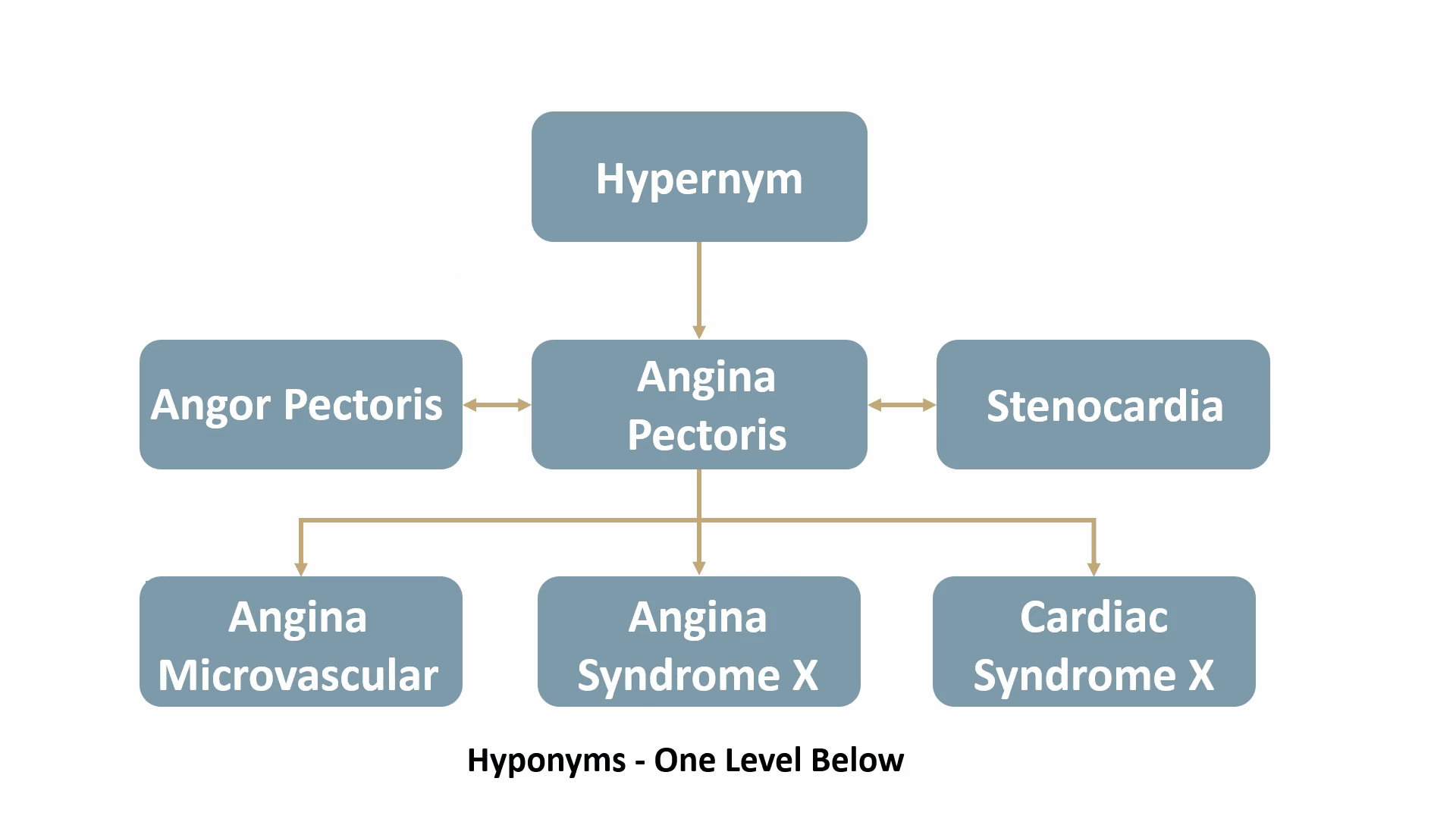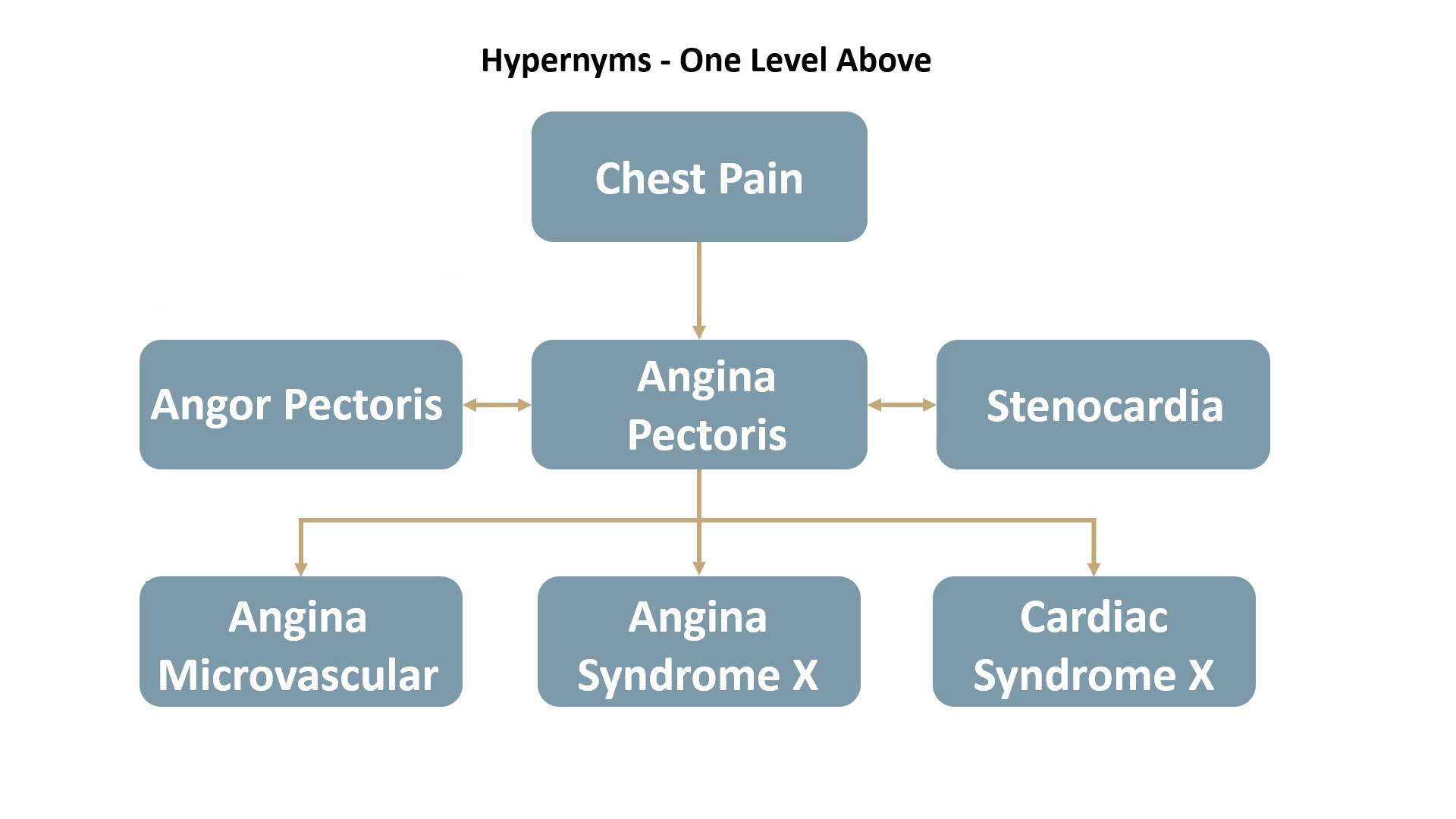
For example, Angina Pectoris, a medical term for chest pain due to coronary heart disease, would be mapped out as shown below:
ICD-10
The International Statistical Classification of Diseases and Related Health Problems (ICD) is a standardized system for coding medical diagnoses and procedures. It contains a description of all known diseases and injuries across languages and allows quick access to disease information, including symptoms, treatment, and mortality rates.

The ICD-10 system has been adopted by numerous countries worldwide and finds use in a wide array of healthcare settings, including hospitals, clinics, and various healthcare facilities. Fully implemented in 2015, ICD-10 boasts an impressive array of over 150,000 codes covering a broad spectrum of medical conditions and treatments. The ICD ontology continues to evolve, and while the next version, ICD-11, is already available for implementation, it is not expected to be fully launched until 2025 or later.
Codes in ICD-10 can have 3 to 7 characters, where the diagnosis category is the first 3 characters. The next set of characters in the code constitutes the related etiology, anatomic site, severity, or other details. The seventh character (if used) specifies any extension classifier describing the episode of care. For example, if a medical record stated that “the patient developed a chalazion of the left upper eyelid,” this would be coded as H00.14 in the ICD-10 coding system.

MedDRA
The Medical Dictionary for Regulatory Activities (MedDRA) is an ontology for classifying and sharing regulatory information. This medical ontology is typically used by pharmaceutical and biotechnology companies for FDA pharmacovigilance reporting on drugs and therapies. MedDRA is used for registration, documentation, and safety monitoring of medical products before and after a product has been authorized for sale. This includes pharmaceuticals vaccines and drug-device combination products.

MedDRA contains over 70,000 standard terms for various signs, symptoms, diseases, diagnoses, and indications, and is updated twice a year. It consists of a 5-level taxonomy, arranging symptoms and diseases from very specific to very general. For example, consider the condition known as “chest pain” (stenocardia). The 5-level hierarchical taxonomy describing this is summarized by the following animation.
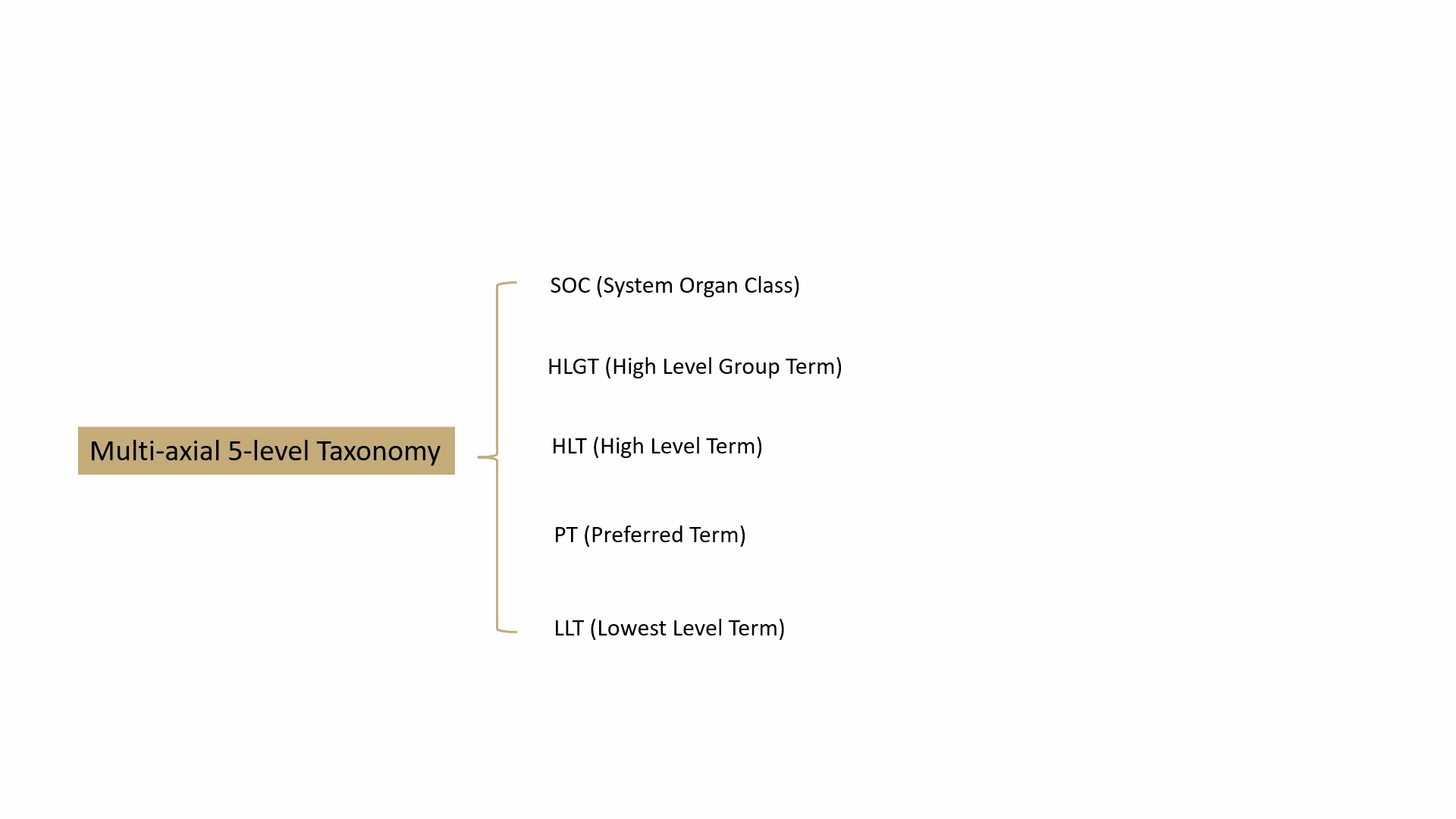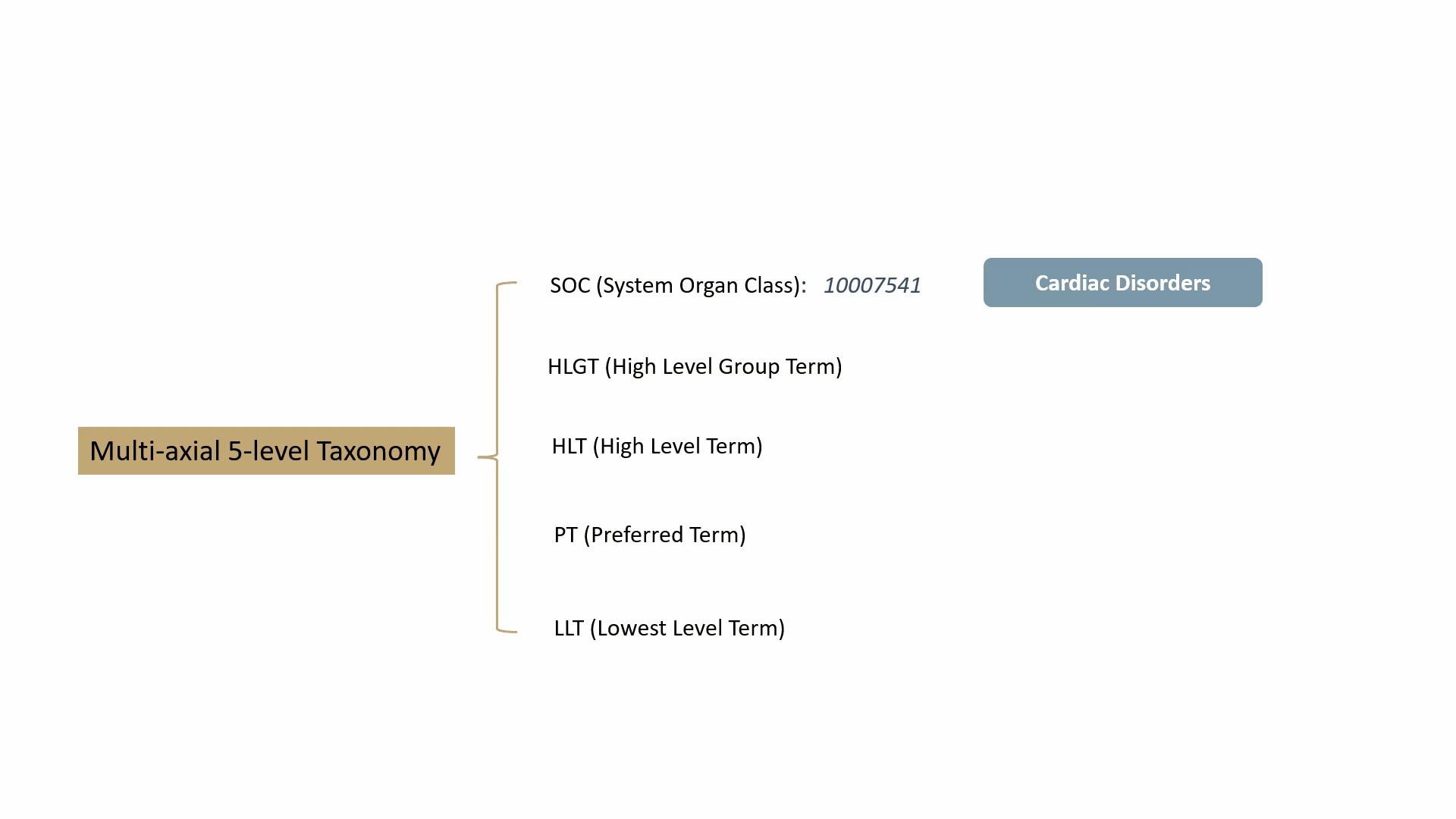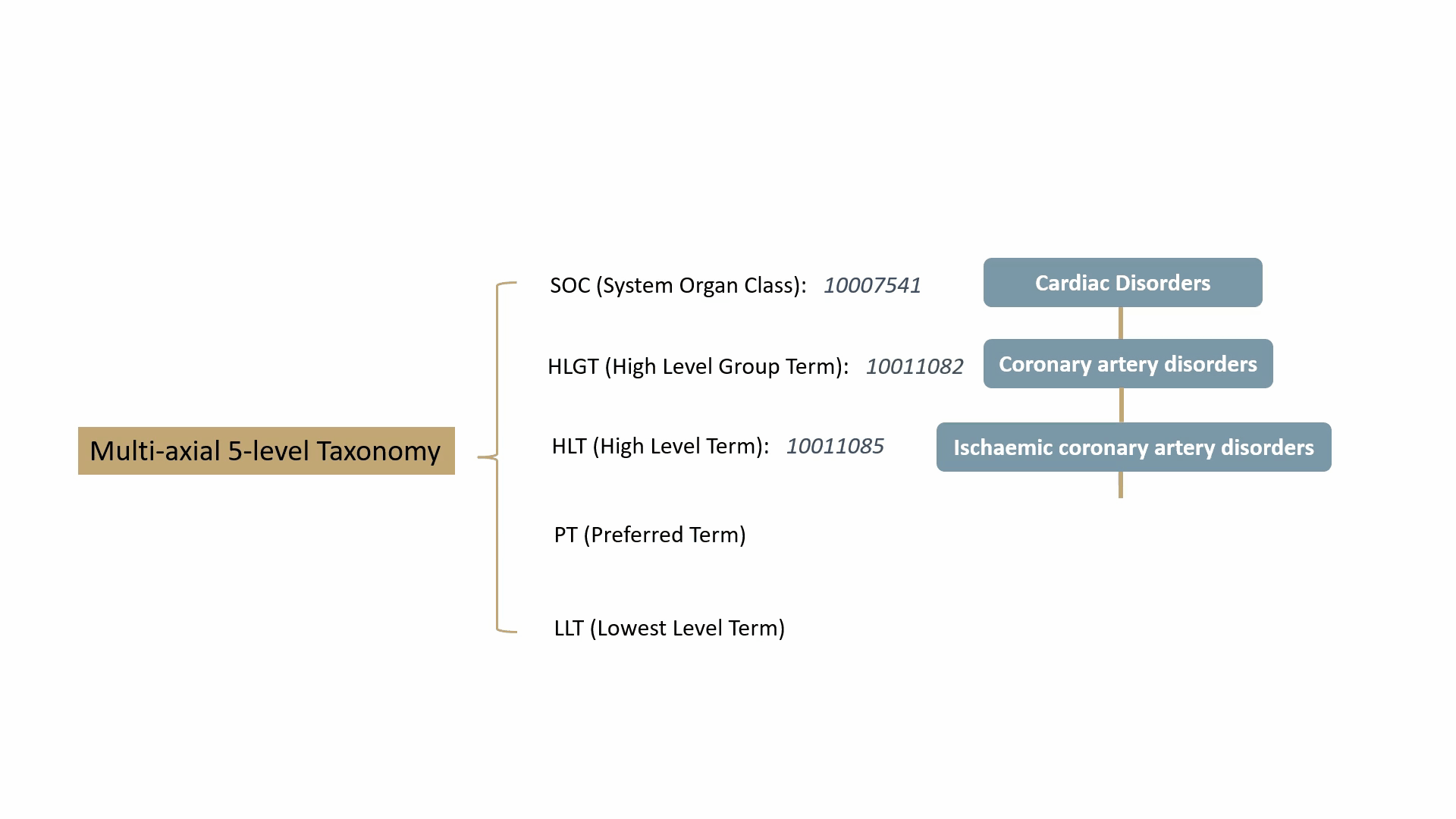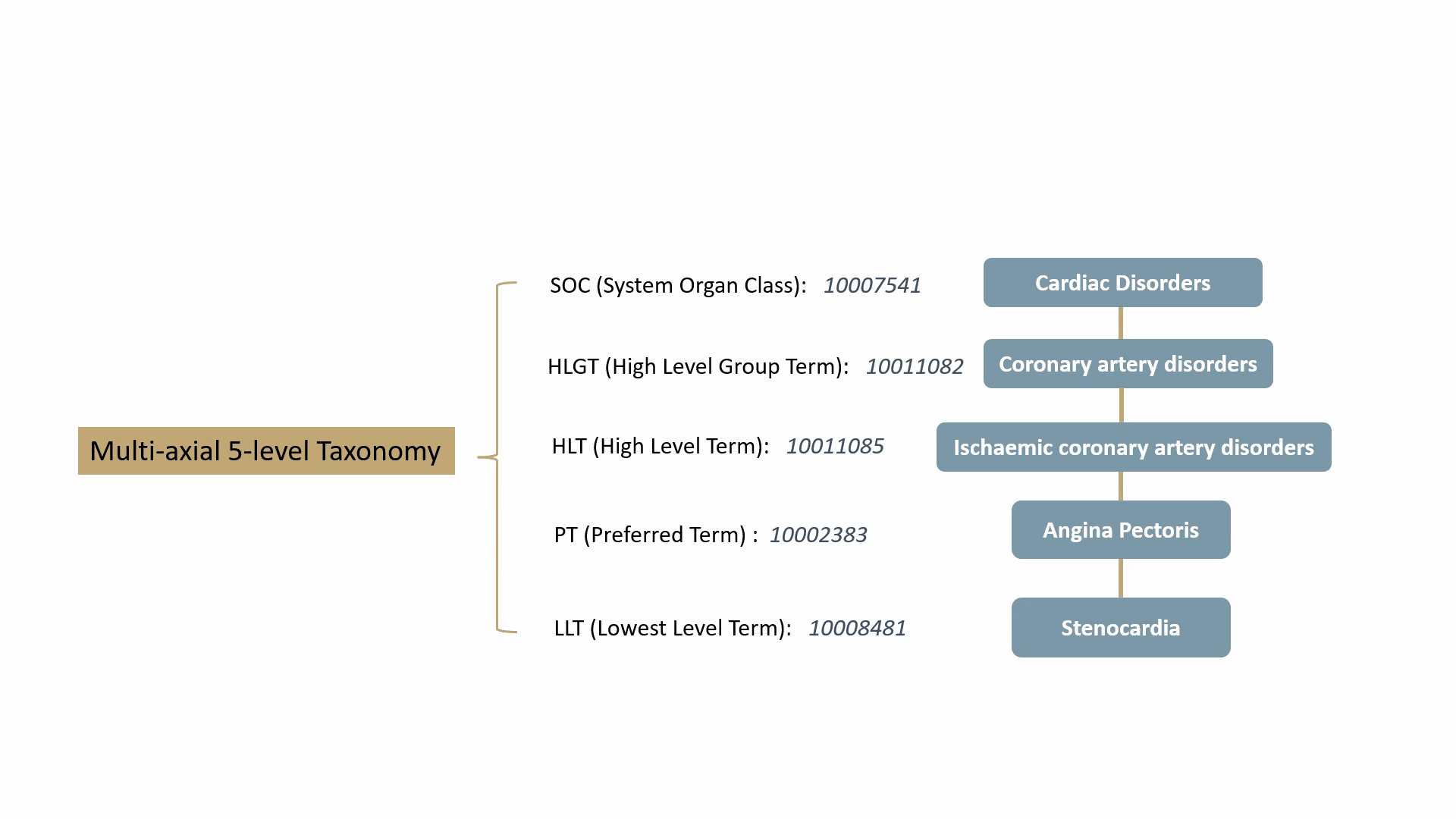
MedDRA is dictated by the FDA in the United States. The FDA requires the use of MedDRA in the submission of safety reports and other regulatory documents related to drug development and approval. It’s widely recognized as a key standard for the classification and analysis of medical data in the pharmaceutical industry.
RxNorm
RxNorm is a comprehensive database containing over 100,000 medications and chemicals. It serves as a standardized nomenclature system that establishes connections between brand names and generic synonyms for clinical drugs. This mapping enables medical care providers to effectively manage the diverse vocabulary associated with drug and pharmaceutical products across various databases.

Moreover, RxNorm’s primary aim is to facilitate efficient and accurate communication of drug-related information among computer systems. It is diligently maintained and updated on a weekly basis by the National Library of Medicine (NLM) and encompasses all medications for humans available on the US market.
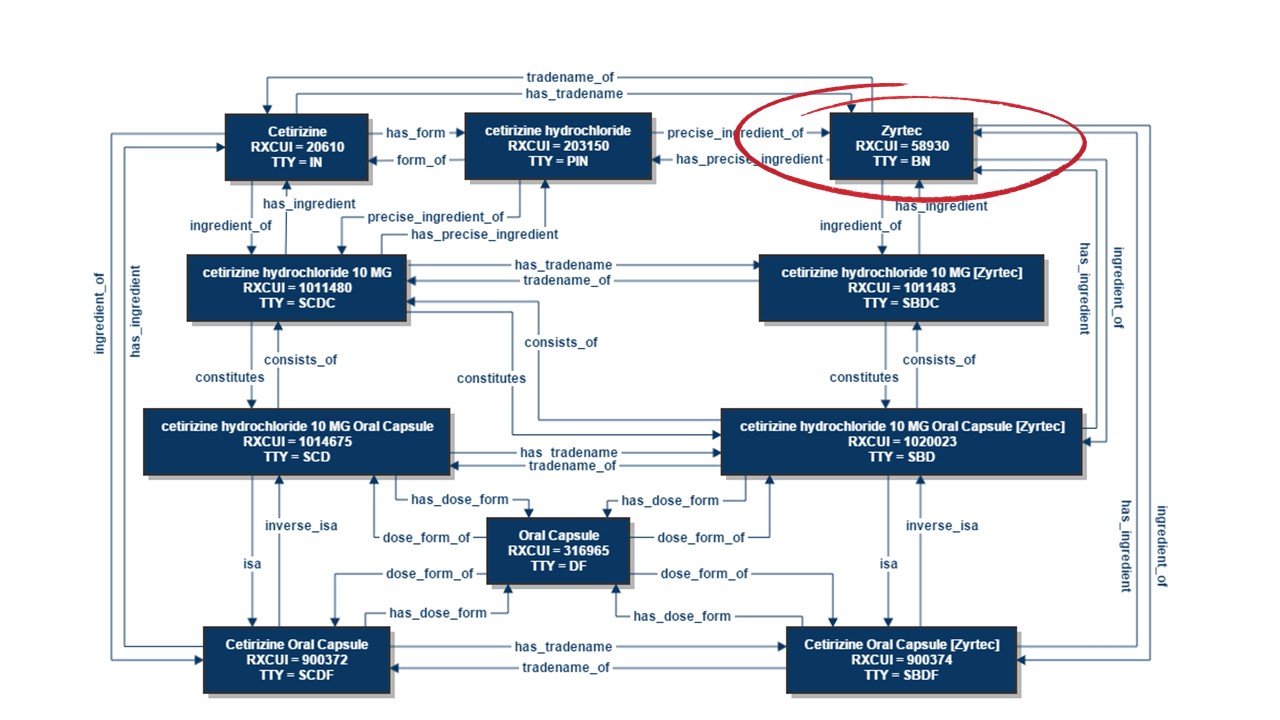
RxNorm serves as an exemplary instance of a highly intricate ontology, as demonstrated in the example below that showcases the connections for Zyrtec® (cetirizine hydrochloride), an over-the-counter antihistamine. This not only provides the standard drug names but also highlights the relationships between different drug entities.
Other sources for ontologies
A few additional sources of coding systems and ontologies include:
- Current Procedural Terminology (CPT) codes: an ontology used to report procedures and diagnostic services to healthcare providers, insurance companies, and accredited institutions.
- Systemized Nomenclature of Medicine – Clinical Terms (SNOMED-CT): an ontology used mainly by healthcare providers to share information in a more standardized manner.
- Diagnosis-Related Group (DRG) codes: a system used to classify hospital cases into approximately 500 groups for the purpose of payment.
- Gene Ontology Consortium: a set of frameworks for modeling complex biological systems and concepts.
How ontologies are used in PolyAnalyst™
PolyAnalyst™ utilizes ontologies to search for and extract pertinent information. Within PolyAnalyst™, ontologies are imported as distinct “semantic dictionaries,” which are then used by PDL (Pattern Definition Language) to extract text patterns that correspond to various medical entities and their relationships as defined within those semantic dictionaries. In addition to the existing ontologies, users have the ability to create their own ontologies.
PolyAnalyst can import and start working with any kinds of ontologies. Some of these ontologies can be used free of charge, others require the user to obtain a license from the ontology owner. Currently, the following ontologies are provided along with PolyAnalyst™ (out-of-the box):
- Human Disease Ontology: provides a framework for organizing and integrating information related to diseases in humans.
- Human Phenotype Ontology: provides a standardized vocabulary of phenotypic abnormalities encountered in human disease.
- MeSH: covers a wide range of topics related to health and medicine.
- RXNorm: contains information on medication names and their active ingredients.
- WordNet: a broad semantic ontology that attempts to build a lexical database of English nouns, verbs, adjectives, and the relationships between them.
Other ontologies can be imported by the user (possibly, after acquiring the necessary license).
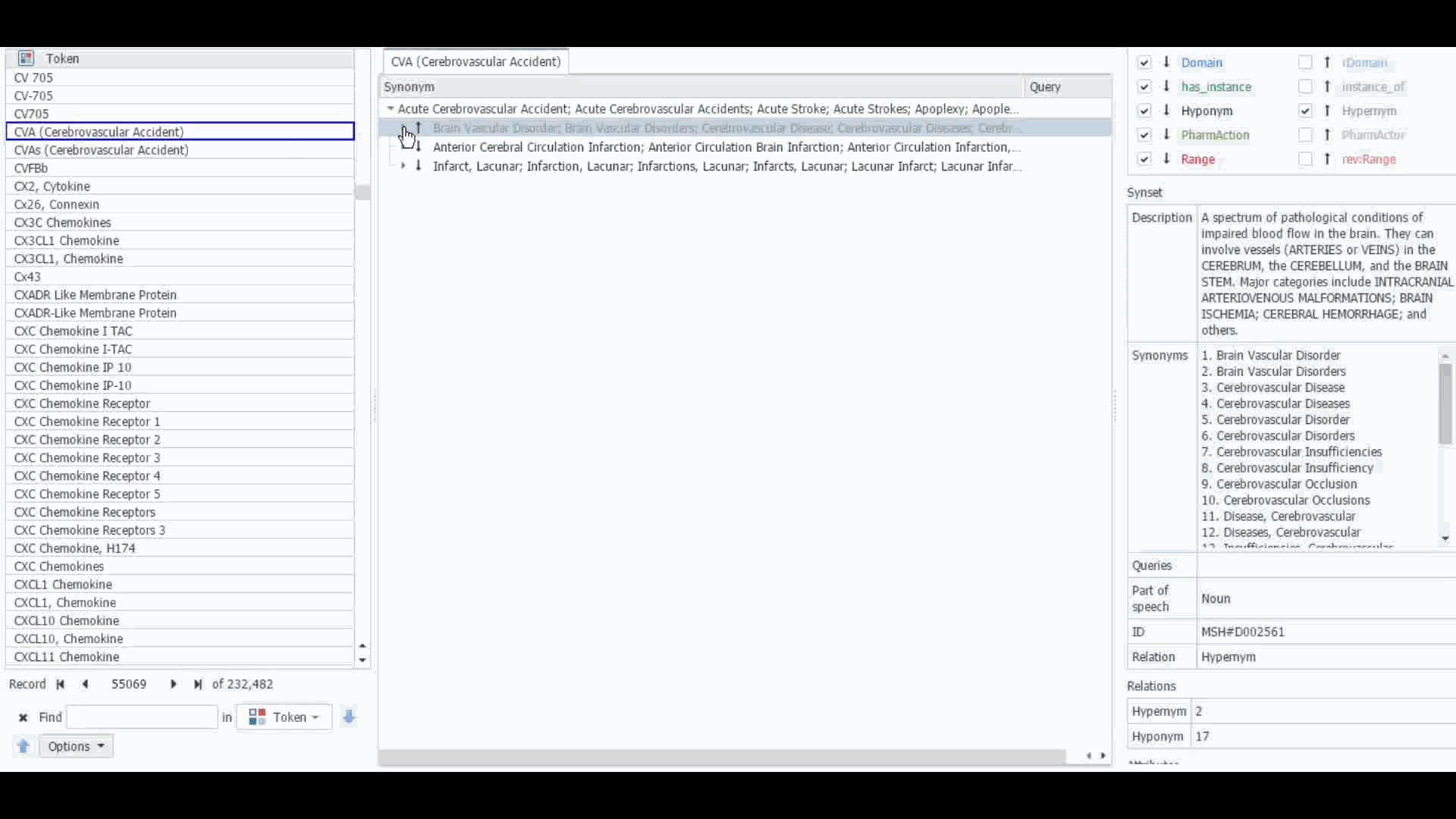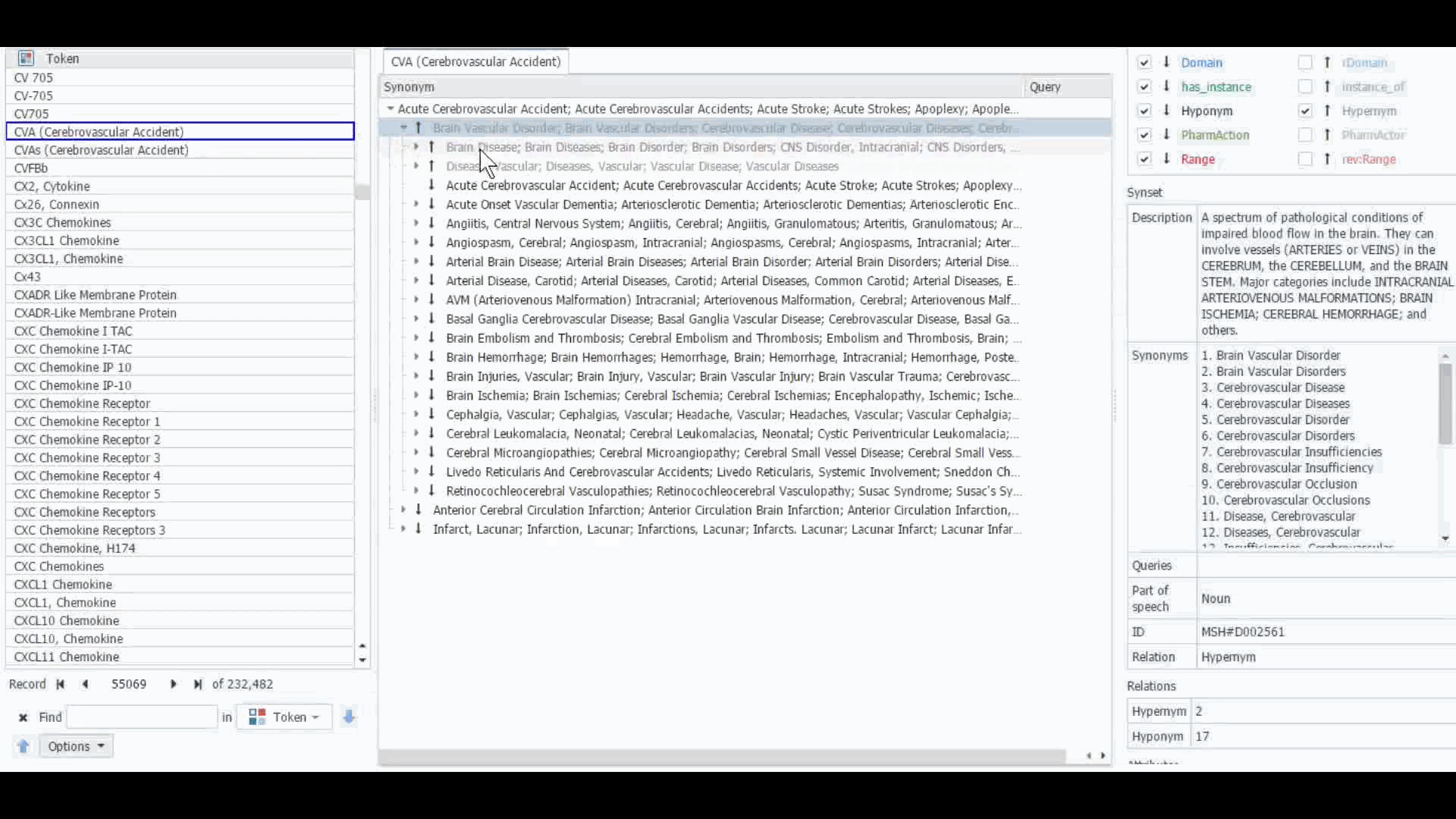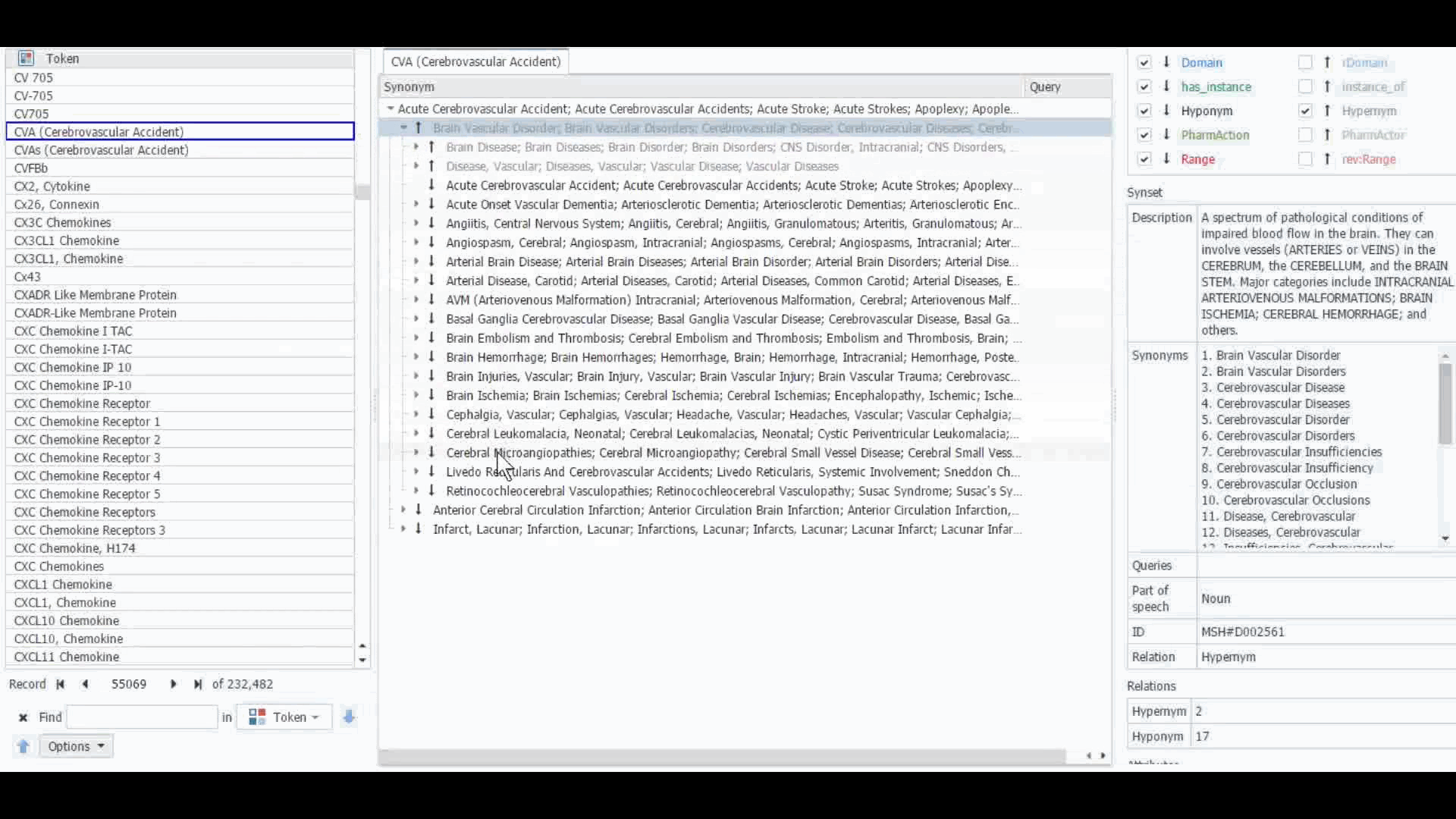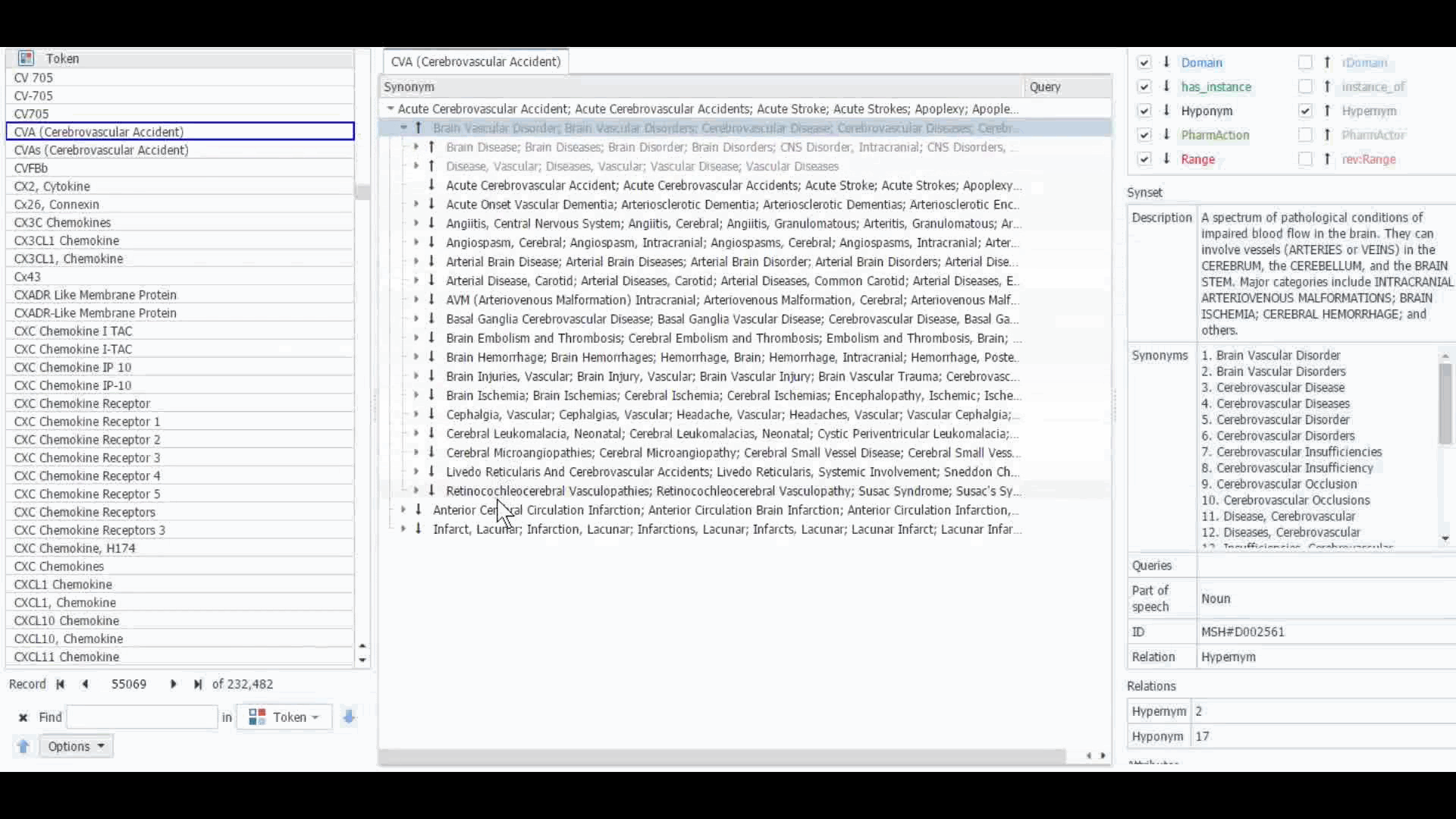
The Dictionary Manager in PolyAnalyst™ is the most direct way to access, edit, and view any imported ontologies in PolyAnalyst™. For example, you can import and view MeSH dictionary in PolyAnalyst™, searching for terms such as “CVA” to view all the associated information.
These ontologies can also be accessed within the project nodes. Some key nodes that utilize these types of ontologies include Entity Extraction and Taxonomy. Within these nodes, users have the option to select which ontology to import. Subsequently, they can craft a query to identify semantically related terms, such as synonyms, hyponyms, or hypernyms, or any other semantic relations, as defined within that particular ontology.
The example below illustrates how a query searching for medical conditions within the “influenza and pneumonia” category, as per ICD-10, can be simplified by leveraging the PDL functions integrated into PolyAnalyst™ in conjunction with the MeSH ontology. By employing the “hyponym” function, as shown below, it becomes possible to capture all the subtypes of influenza and pneumonia without the need to list them individually.
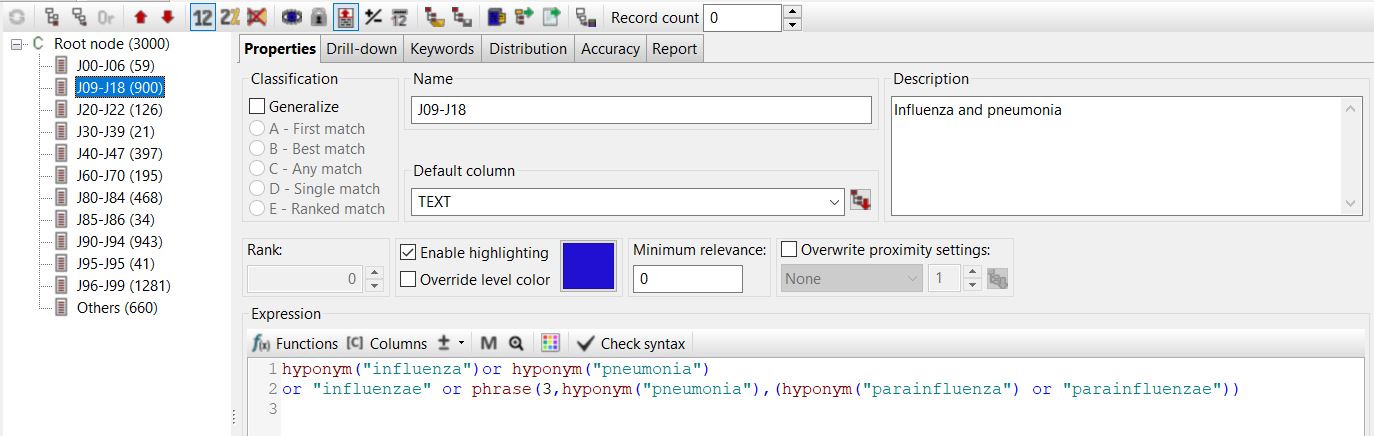
Lastly, one of the challenges of using ontologies is the issue of ontology cleaning, which involves removing or correcting errors, inconsistencies and irrelevant or outdated information in the ontology. This is particularly important in specialized domains, such as chemistry or toxicology, where the ontology may contain a large number of concepts and entities that are not relevant to the domain or may cause confusion or misinterpretation (i.e., “milk” or “coffee” being present within a chemical substance ontology). Thankfully, the task of modifying ontologies is quite straight-forward within the Dictionaries module of PolyAnalyst™.
Ontologies and beyond
Ontologies are powerful tools in information science, with potential that far exceeds the medical domain of the topics discussed here. Additional resources on using dictionaries in PolyAnalyst™ and how to import and utilize ontologies and specialized dictionaries can be found in our support and training materials.
