When we begin a search on the internet, we often use a simple keyword to find exactly what we need. With specified information captured, there are several backend components that feed into the end result. Naturally, we take for granted this instantaneous retrieval. What goes on behind that search contributes to its relevancy, such as the context, the scope, and whether to include information adjacently related to the keyword. Similar to a simple search on the internet, we can recapture information from large unstructured text data by using the same process.
For example, if we wanted to collect a subset of medical records associated with cancer, we could search the database for the word ‘cancer’. This simple search would return plenty of records containing this literal string:
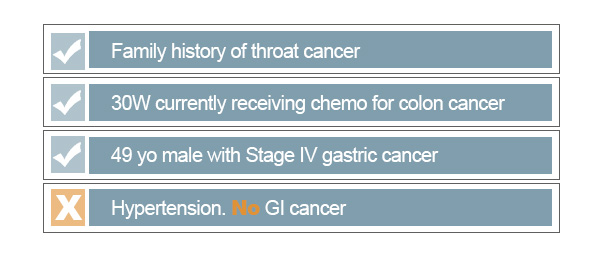
 As shown above, there are some records that may not be good matches for what we intend for the cancer subset. The first thing that we need to ensure is that we avoid returning negations of a keyword. For this search, we would certainly not like to include records in which there are no signs of cancer. Searching for a keyword alone fails to recognize linguistic patterns around it, such as negation. By restricting our query to filter out false positives of cancer, we can significantly improve accuracy of our search. PolyAnalyst™ has built-in features that innately recognize complex signs of negation to automate such filtering.
As shown above, there are some records that may not be good matches for what we intend for the cancer subset. The first thing that we need to ensure is that we avoid returning negations of a keyword. For this search, we would certainly not like to include records in which there are no signs of cancer. Searching for a keyword alone fails to recognize linguistic patterns around it, such as negation. By restricting our query to filter out false positives of cancer, we can significantly improve accuracy of our search. PolyAnalyst™ has built-in features that innately recognize complex signs of negation to automate such filtering.
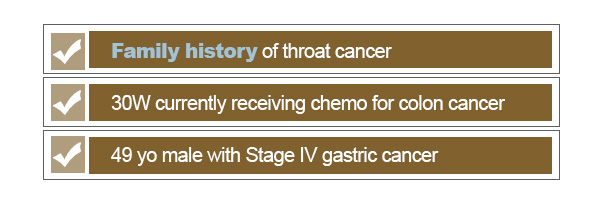
 Another consideration is the intention or context of the search. When we are looking for records that are related to cancer, are we looking for patients who have developed cancer? Do we want to ignore family history of cancer or cancer risks associated with a patient’s medications? The breadth of a search is largely dependent on the types of questions that we are looking to address with the search.
Another consideration is the intention or context of the search. When we are looking for records that are related to cancer, are we looking for patients who have developed cancer? Do we want to ignore family history of cancer or cancer risks associated with a patient’s medications? The breadth of a search is largely dependent on the types of questions that we are looking to address with the search.
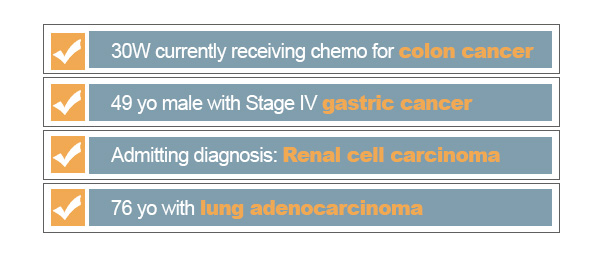
 Of course, the search gets even more complex when we consider the many ways to express cancer as well as its symptoms. We may be missing true matches of cancer related records because we are not capturing certain pathology names, synonyms, symptoms, and other terms related to this condition. One could try to search for every technical term associated with a specific keyword, but that would be needlessly inefficient and would require a high level of domain knowledge. For the pharmaceutical and medical domains, a professional may even reference trusted knowledge bases such as MeSH and MeDRA to study terms semantically related to the medical keyword of interest. Instead of compiling this knowledge into every query, it makes sense to automate this process with a semantic ontology to not only get returns for ‘cancer’ but also synonyms and subtypes of cancer. Further, one could even expand this search by including cancer symptoms and medications used to treat cancers.
Of course, the search gets even more complex when we consider the many ways to express cancer as well as its symptoms. We may be missing true matches of cancer related records because we are not capturing certain pathology names, synonyms, symptoms, and other terms related to this condition. One could try to search for every technical term associated with a specific keyword, but that would be needlessly inefficient and would require a high level of domain knowledge. For the pharmaceutical and medical domains, a professional may even reference trusted knowledge bases such as MeSH and MeDRA to study terms semantically related to the medical keyword of interest. Instead of compiling this knowledge into every query, it makes sense to automate this process with a semantic ontology to not only get returns for ‘cancer’ but also synonyms and subtypes of cancer. Further, one could even expand this search by including cancer symptoms and medications used to treat cancers.
 With these considerations, it becomes easier to search smarter and extract exactly what you need from your unstructured text. Utilizing the effectiveness of simple keyword searches combined with the analysis of linguistic patterns and semantic relations, the automated querying of unstructured data returns the sought relevant information in a tiny fraction of the processing time compared to manual searches.
With these considerations, it becomes easier to search smarter and extract exactly what you need from your unstructured text. Utilizing the effectiveness of simple keyword searches combined with the analysis of linguistic patterns and semantic relations, the automated querying of unstructured data returns the sought relevant information in a tiny fraction of the processing time compared to manual searches.
