We have electronic medical records containing free text descriptions as well as structured information like patient IDs, hospital admission IDs, and date/time. One patient may have multiple additions to his or her medical record over the course of one hospital stay, so there may be several records with the same patient ID and hospital admission ID, but different notes and timestamps.
The Problem
We want to use free text in medical records to predict when a patient will develop sepsis. We know which patients were eventually billed for a sepsis diagnosis, but we don’t want to include notes that were written after the patient developed sepsis, or we might discover useless predictors like “began course of antibiotics for treatment of sepsis”. Once sepsis is suspected, the data is no longer useful for early prediction. How can we remove the records that mention sepsis, as well as all the patient’s other records with later timestamps, even if the later notes do not directly mention sepsis?
The Plan
We are going to solve this problem using PolyAnalyst, Megaputer’s data processing software. The following is a screenshot of a flowchart built using the software. The icons represent individual processing steps. Each icon is described below.
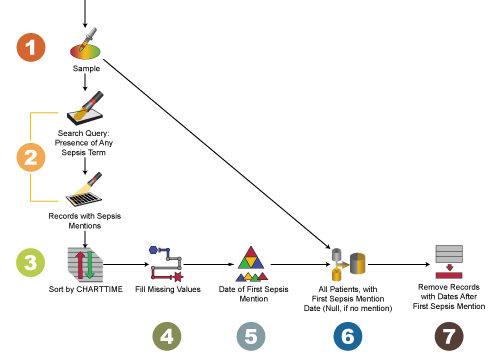
- Begin with a dataset of electronic medical records. I’ll use a small sample for testing my workflow first. Node: Sample.
- Search for mentions of sepsis in the text, and get a subset containing just those records. These will serve as the cutoff times for the patients they represent. Remember, we want to keep only the notes that the patient received beforethis time. Node: Search Query and Search Query Subset. The latter can be created by right-clicking on the relevant query in the Search Query node and selecting “Create subset”.
- Sort the records by time: earliest dates first. Node: Sort Rows.
- Create a new column for timestamp information in each row, called “Earliest Sepsis Mention”. In our case, some timestamp entries were empty and needed to be filled with a more general date. Node: Derive.
- For any patient ID / hospital admission ID pair, keep only the first record. You now have a list of patients, each with a value in a column called “Earliest Sepsis Mention,” containing the timestamp for the first record that mentions their sepsis. Node: Aggregate.
- Join this dataset (right) with your original dataset (left). Use a left outer join so that for every record in your original dataset, it checks your new list to see if there is a matching patient and hospital admission ID. If there is, it will add in the new information: the timestamp of that patient’s Earliest Sepsis Mention. If that patient and hospital admission are not in the new list (they never had sepsis mentioned in their notes), the Earliest Sepsis Mention will remain blank. Node: Join.
- Now for the easy part! Filter rows so that you only keep the records with an empty value in Earliest Sepsis Mention, or where the record’s timestamp is an earlier time than that patient’s Earliest Sepsis Mention. Node: Filter Rows.
The Result
We searched our data for mentions of sepsis, selected the earliest record for each patient in that list, then applied that answer to all patient records in our original dataset. From there, it was easy to compare the medical note timestamp with the timestamp of the first time sepsis was ever mentioned for that patient, meaning we have a new, smaller dataset where we have eliminated all of the records mentioning sepsis, as well as those patients’ later follow-up records.
In short, we flagged all patient records with timestamps after a particular event in their medical record, so they could be removed from our training data.
